Notes on Multivariate Data Analysis via Matrix Decomposition
2019-09-30
These are the notes taken on my master course Multivariate Data Analysis via Matrix Decomposition. If you’re confused with the course name, you can think of this as a statistical course on unsupervised learning.
$\newcommand{1}[1]{\unicode{x1D7D9}_{\{#1\}}}\newcommand{Corr}{\text{Corr}}\newcommand{E}{\text{E}}\newcommand{Cov}{\text{Cov}}\newcommand{Var}{\text{Var}}\newcommand{span}{\text{span}}\newcommand{bs}{\boldsymbol}\newcommand{R}{\mathbb{R}}\newcommand{rank}{\text{rank}}\newcommand{\norm}[1]{\left\lVert#1\right\rVert}\newcommand{diag}{\text{diag}}\newcommand{tr}{\text{tr}}\newcommand{braket}[1]{\left\langle#1\right\rangle}\newcommand{C}{\mathbb{C}}$
Notations
First let’s give some standard notations used in this course. Let’s assume no prior knowledge in linear algebra and start from matrix multiplication.
Matrix Multiplication
We denote a matrix $\mathbf{A}\in\R^{m\times n}$, with its entries defined as $[a_{ij}]_{i,j=1}^{m,n}$. Similarly, we define $\mathbf{B}=[b_{jk}]_{j,k=1}^{n,p}$ and thus the multiplication is defined as $\mathbf{AB} = [\sum_{j=1}^n a_{ij}b_{jk}]_{i,k=1}^{n,p}$, which can also be represented in three other ways:
- vector form, using
$\mathbf{a}$and$\mathbf{b}$ - a matrix of products of
$A$and$\mathbf{b}$ - a matrix of products of
$\mathbf{a}$and$\mathbf{B}$
A special example of such representation: let’s assume
$$ \mathbf{A}=[\mathbf{a}_1,\mathbf{a}_2,\ldots,\mathbf{a}_n]\in\R^{m\times n}\text{ and } \mathbf{D} = \diag(d_1,d_2,\ldots,d_n) \in\R^{n\times n},$$
then we have right away $\mathbf{AD}=[\mathbf{a}_id_i]_{i=1}^n$.
Exercise With multiplication we care ranks of matrices. There is a quick conclusion: If $\mathbf{x}\neq \bs{0}, \mathbf{y}\neq \bs{0}$, then $\rank(\bs{yx'})=1$. Conversely, if $\rank(\mathbf{A})=1$, then $\exists\ \mathbf{x}\neq \bs{0}, \mathbf{y}\neq \bs{0}$ s.t. $\bs{xy'}=\mathbf{A}$. Prove it.
Norms
There are two types of norms in this course we consider:
- (Euclidean) We define the
$l^1$-norm as$\norm{\mathbf{x}}_1 = \sum_{i=1}^n |x_i|$, define$l^2$-norm as$\norm{\mathbf{x}}_2 = \sqrt{\mathbf{x'x}}$, define$l^{\infty}$-norm as$\norm{\mathbf{x}}_{\infty} = \max_{1\le i \le n}\{|x_i|\}$, and define the Mahalanobis norm as$\norm{\mathbf{x}}_A = \sqrt{\mathbf{x'Ax}}$. - (Frobenius) We define the Frobenius norm of a matrix as
$\norm{\mathbf{A}}_F=\sqrt{\sum_{i=1}^m\sum_{j=1}^n a_{ij}^2}$. The spectral 2-norm of a matrix is defined as$\norm{\mathbf{A}}_2=\max_{\mathbf{x}\neq \bs{0}} \norm{\mathbf{Ax}}_2 / \norm{\mathbf{x}}_2$.
Properties of these norms:
$\norm{\mathbf{v}}=0$iff.$\mathbf{v}=\bs{0}$.$\norm{\alpha \mathbf{v}} = |\alpha|\cdot\norm{\mathbf{v}}$for any$\alpha\in\R$and any$\mathbf{v}\in\mathcal{V}$.- (Triangular Inequality)
$\norm{\mathbf{u} + \mathbf{v}} \le \norm{\mathbf{u}} + \norm{\mathbf{v}}$for any$\mathbf{u}, \mathbf{v}\in\mathcal{V}$. - (Submultiplicative)
$\norm{\mathbf{AB}}\le \norm{\mathbf{A}}\cdot \norm{\mathbf{B}}$for every formable matrices$\mathbf{A}$and$\mathbf{B}$.
Exercise Try to prove them for Euclidean 2-norm, Frobenius norm and spectral 2-norm.
Inner Products
There are two types of inner products we consider:
- (Euclidean) We define the inner product of vectors
$\mathbf{x},\mathbf{y}\in\R^n$as$\mathbf{x'y}=\sum_{i=1}^n x_iy_i$. - (Frobenius) We define the inner product of matrices
$\mathbf{A},\mathbf{B}\in\R^{m\times n}$as$\braket{\mathbf{A},\mathbf{B}}=\tr(\mathbf{A'B})=\sum_{i=1}^m\sum_{j=1}^n a_{ij}b_{ij}$.
A famous inequality related to these inner products is the Cauchy-Schwarz inequality, which states
- (Euclidean)
$|\mathbf{x'y}|\le \norm{\mathbf{x}}_2\cdot\norm{\mathbf{y}}_2$for any$\bs{x,y}\in\R^n$. - (Frobenius)
$|\braket{\mathbf{A},\mathbf{B}}|\le\norm{\mathbf{A}}_F\cdot\norm{\mathbf{B}}_F$for any$\mathbf{A},\mathbf{B}\in\R^{m\times n}$.
Eigenvalue Decomposition (EVD)
The first matrix decomposition we’re gonna talk about is the eigenvalue decomposition.
Eigenvalues and Eigenvectors
For square matrix $\mathbf{A}\in\R^{n\times n}$, if $\bs{0}\neq \mathbf{x}\in\C^n$ and $\lambda\in\C$ is s.t. $\mathbf{Ax} = \lambda\mathbf{x}$, then $\lambda$ is called an engenvalue of $\mathbf{A}$ and $\mathbf{x}$ is called the $\lambda$-engenvector of $\mathbf{A}$.
Ideally, we want a matrix to have $n$ eigenvectors and $n$ corresponding eigenvectors, linearly independent to each other. This is not always true.
Existence of EVD
Theorem $\mathbf{A}\in\R^{n\times n}$ have $n$ eigenvalues iff. there exists an invertible $\mathbf{X}\in\R^{n\times n}$ s.t. $\mathbf{X}^{-1}\mathbf{A}\mathbf{X}=\bs{\Lambda}$, i.e. $\mathbf{A}$ is diagonizable. This gives $\mathbf{A}=\mathbf{X}\bs{\Lambda}\mathbf{X}^{-1}$, which is called the eigenvalue decomposition (EVD).
Theorem (Spectral Theorem for Symmetric Matrices) For symmetric matrix $\mathbf{A}\in\R^{n\times n}$ there always exists an orthogonal matrix $\mathbf{Q}$, namely $\mathbf{Q}'\mathbf{Q}=\mathbf{I}$, that gives
$$ \mathbf{A}=\mathbf{Q}\bs{\Lambda}\mathbf{Q}’ = \sum_{i=1}^n \lambda_i \mathbf{q}_i \mathbf{q}_i' $$
where $\mathbf{q}$ are column vectors of $\mathbf{Q}$. This is called the symmetric EVD, aka. $\mathbf{A}$ being orthogonally diagonalizable.
Properties of EVD
We have several properties following the second theorem above. For all $i=1,2,\ldots, n$
$\mathbf{A}\mathbf{q}_i = \lambda_i \mathbf{q}_i$(can be proved using$\mathbf{Q}^{-1}=\mathbf{Q}'$)$\norm{\mathbf{q}_i}_2=1$(can be proved using$\mathbf{QQ}'=\mathbf{I}$)
The second theorem above can also be represented as
Theorem If $\mathbf{A}=\mathbf{A}'$, then $\mathbf{A}$ has $n$ orthogonal eigenvectors.
Singular Value Decomposition (SVD)
For general matrices, we have singular value decomposition.
Definition
The most famous form of SVD is define as
$$ \mathbf{A} = \mathbf{U} \bs{\Sigma} \mathbf{V}' $$
where $\mathbf{A}\in\R^{m\times n}$, $\mathbf{U}\in\R^{m\times m}$, $\bs{\Sigma}\in\R^{m\times n}$ and $\mathbf{V}\in\R^{n\times n}$. Specifically, both $\mathbf{U}$ and $\mathbf{V}$ are orthogonal (i.e. $\mathbf{U}'\mathbf{U}=\mathbf{I}$, same for $\mathbf{V}$) and $\bs{\Sigma}$ is diagonal. Usually, we choose the singular values to be non-decreasing, namely
$$ \bs{\Sigma}=\diag(\sigma_1,\sigma_2,\ldots,\sigma_{\min{m,n}})\quad\text{where}\quad \sigma_1 \ge \sigma_2 \ge \cdots \ge \sigma_{\min{m,n}}. $$
Terminology
Here we define a list of terms that’ll be used from time to time:
- (SVD)
$\mathbf{A} = \mathbf{U} \bs{\Sigma} \mathbf{V}'$. - (Left Singular Vectors) Columns of
$\mathbf{U}$. - (Right Singular Vectors) Columns of
$\mathbf{V}$. - (Singular Values) Diagonal entries of
$\bs{\Sigma}$.
Three Forms of SVD
Besides the regular SVD given above, we have the outer product SVD:
$$ \mathbf{A} = \sum_{i=1}^{\min{m,n}}\mkern-10mu\sigma_i \mathbf{u}_i \mathbf{v}_i' $$
and condensed SVD:
$$ \mathbf{A} = \mathbf{U}_r\bs{\Sigma}_r\mathbf{V}_r' $$
where $r=\rank(\mathbf{A})$ is also the number of non-zero singular values. In this form, we have $\bs{\Sigma}_r\in\R^{r\times r}$ with proper chunked $\mathbf{U}_r$ and $\mathbf{V}_r$.
Existence of SVD
Theorem (Existence of SVD) Let $\mathbf{A}\in\R^{m\times n}$ and $r=\rank(\mathbf{A})$. Then $\exists\ \mathbf{U}_r\in\R^{m\times r}$, $\mathbf{V}_r\in\R^{n\times r}$ and $\bs{\Sigma}_r\in\R^{r\times r}$ s.t. $\mathbf{A} = \mathbf{U}_r\bs{\Sigma}_r\mathbf{V}_r'$ where $\mathbf{U}_r$ and $\mathbf{V}_r$ are orthogonal and $\bs{\Sigma}_r$ is diagonal. This means condensed SVD exists and therefore the rest two forms.
Proof. Define symmetric $\mathbf{W}\in\R^{(m+n)\times(m+n)}$ as
$$ \mathbf{W} = \begin{bmatrix} \bs{0} & \mathbf{A} \ \mathbf{A}’ & \bs{0} \end{bmatrix} $$
which has an orthogonal EVD as $\mathbf{W} = \mathbf{Z}\bs{\Lambda}\mathbf{Z}'$ where $\mathbf{Z}'\mathbf{Z}=\mathbf{I}$. Now, assume $\mathbf{z}\in\R^{m+n}$ is an eigenvector of $\mathbf{W}$ corresponding to $\lambda$, then $\mathbf{W}\mathbf{z} = \lambda \mathbf{z}$. Denote the first $m$ entries of $\mathbf{z}$ as $\mathbf{x}$ and the rest $\mathbf{y}$, which gives
$$ \begin{bmatrix} \bs{0} & \mathbf{A}\ \mathbf{A}’ & \bs{0} \end{bmatrix} \begin{bmatrix} \mathbf{x} \ \mathbf{y} \end{bmatrix} = \lambda \begin{bmatrix} \mathbf{x} \ \mathbf{y} \end{bmatrix} \Rightarrow \begin{cases} \mathbf{Ay} = \lambda \mathbf{x},\ \mathbf{A}’\mathbf{x} = \lambda \mathbf{y}. \end{cases} $$
Using this results
$$ \begin{bmatrix} \bs{0} & \mathbf{A}\ \mathbf{A}’ & \bs{0} \end{bmatrix} \begin{bmatrix} \mathbf{x} \ -\mathbf{y} \end{bmatrix} = \begin{bmatrix} -\mathbf{Ay} \ \mathbf{A}’\mathbf{y} \end{bmatrix} = \begin{bmatrix} -\lambda \mathbf{x}\ \lambda \mathbf{y} \end{bmatrix} = -\lambda\begin{bmatrix} \mathbf{x}\ -\mathbf{y} \end{bmatrix} $$
which means $-\lambda$ is also an engenvalue of $\mathbf{W}$. Hence, we know
$$ \begin{align} \mathbf{W} &= \mathbf{Z}\bs{\Lambda}\mathbf{Z}’ = \mathbf{Z}_r\bs{\Lambda}_r\mathbf{Z}_r’\ &= \begin{bmatrix} \mathbf{X} & \mathbf{X}\ \mathbf{Y} & -\mathbf{Y} \end{bmatrix} \begin{bmatrix} \bs{\Sigma} & \bs{0}\ \bs{0} & -\bs{\Sigma} \end{bmatrix} \begin{bmatrix} \mathbf{X} & \mathbf{X}\ \mathbf{Y} & -\mathbf{Y} \end{bmatrix}’\ &= \begin{bmatrix} \bs{0} & \mathbf{X}\bs{\Sigma}\mathbf{Y}’\ \mathbf{Y}\bs{\Sigma}\mathbf{X}’ & \bs{0} \end{bmatrix}. \end{align} $$
Therefore, we conclude $\mathbf{A}=\mathbf{X}\bs{\Sigma}\mathbf{Y}'$ where now all we need to prove is the orthogonality of $\mathbf{X}$ and $\mathbf{Y}$. Let’s take a look at $\mathbf{z}=(\mathbf{x},\mathbf{y})$ we just defined. Let
$$ \norm{\mathbf{z}}=\mathbf{z}’\mathbf{z}=\mathbf{x}’\mathbf{x} + \mathbf{y}’\mathbf{y} = 2. $$
From orthogonality of eigenvectors corresponding to different eigenvalues, we also know
$$ \mathbf{z}’\bar{\mathbf{z}} = \mathbf{x}’\mathbf{x} - \mathbf{y}’\mathbf{y} = 0 $$
which altogether gives $\norm{\mathbf{x}}=\norm{\mathbf{y}}=1$.Q.E.D.
How to Calculate SVD
Three steps to calculate the SVD of $\mathbf{A}\in\R^{m\times n}$:
$\bs{\Sigma}$: calculate eigenvalues of$\mathbf{AA}'$, then let$\bs{\Sigma}=\diag\{\sigma_1,\sigma_2,\ldots,\sigma_r\}\in\R^{m\times n}$.$\mathbf{U}$: calculate eigenvectors of$\mathbf{AA}'$, then normalize them to norm$1$, then$\mathbf{U}=(\bs{u_1},\bs{u_2},\ldots,\mathbf{u}_m)$.$\mathbf{V}$: calculate eigenvectors of$\mathbf{A}'\mathbf{A}$, then normalize them to norm$1$, then$\mathbf{V}=(\mathbf{v}_1,\mathbf{v}_2,\ldots,\mathbf{v}_n)$.
Remark: Alternatively you may use formula $\mathbf{U}=\mathbf{AV}\bs{\Sigma}^{-1}\Rightarrow \mathbf{u}_i=\mathbf{Av}_i/\sigma_i$.
Properties of SVD
There are several characteristics we have about SVD:
- The
$\mathbf{W}$decomposition above. - The left singular vector
$\mathbf{v}$given by$\mathbf{Au} = \sigma \mathbf{v}$, and the right singular vector$\mathbf{u}$given by$\mathbf{A}'\mathbf{v} = \sigma \mathbf{u}$. - Relationship with eigenvectors/eigenvalues…
- of
$\mathbf{A}'\mathbf{A}$:$\mathbf{A}'\mathbf{A}\mathbf{u} = \sigma\mathbf{A}'\mathbf{v} = \sigma^2\mathbf{u}$. - of
$\mathbf{AA}'$:$\mathbf{AA}'\mathbf{v} = \sigma\mathbf{A}\mathbf{u} = \sigma^2\mathbf{v}$.
- of
- Frobenius norms (eigenvalues cannot define a norm!):
$\norm{\mathbf{A}}_F^2 = \sum_{i,j=1}^{m,n}a_{ij}^2=\sum_{i=1}^r\sigma_i^2$.$\norm{\mathbf{A}}_2 = \max_{\mathbf{x}\neq 0} \norm{\mathbf{Ax}}_2 / \norm{\mathbf{x}}_2 = \sigma_1$.
Exercise Show how to use SVD to calculate these two norms.
Applications of SVD
1) Projections. One of the most importance usages of SVD is computing projections. $\mathbf{P}\in\R^{n\times n}$ is a projection matrix iff. $\mathbf{P}^2=\mathbf{P}$. More commonly, we consider orthogonal projection $\mathbf{P}$ that’s also symmetric. Now let’s consider dataset $\mathbf{A}\in\R^{m\times n}$ where we have $n$ observations, each with $m$ dimensions. Suppose we want to project this dataset onto $\mathbf{W}\subseteq\R^m$ that has $k$ dimensions, i.e.
$$ \mathbf{W} = \span{\mathbf{q}_1,\mathbf{q}_2,\ldots,\mathbf{q}_k},\quad \mathbf{q}_i’\mathbf{q}_j=\1{i=j} $$
then the projection matrix would be $\mathbf{P}_{\mathbf{W}}=\mathbf{Q}_k\mathbf{Q}_k'$.
Nearest Orthogonal Matrix. The nearest orthogonal matrix of $\mathbf{A}\in\R^{p\times p}$ is given by
$$ \min_{\mathbf{X}’\mathbf{X}=\mathbf{I}}\norm{\mathbf{A}-\mathbf{X}}_F $$
which solves if we have optima for
Now we try to solve
$$ \max_{\mathbf{X}’\mathbf{X}=\mathbf{I}} \tr(\mathbf{A}’\mathbf{X}) $$
and claim the solution is given by $\mathbf{X} = \mathbf{U}\mathbf{V}'$ where $\mathbf{U}$ and $\mathbf{V}$ are derived from SVD of $\mathbf{A}$, namely $\mathbf{A} = \mathbf{U}\bs{\Sigma} \mathbf{V}'$. Proof: We know
$$ \tr(\mathbf{A}’\mathbf{X}) = \tr(\mathbf{V}\bs{\Sigma}’\mathbf{U}’\mathbf{X}) = \tr(\bs{\Sigma}’\mathbf{U}’\mathbf{X}\mathbf{V}) =: \tr(\bs{\Sigma}’\mathbf{Z}) $$
where we define $\mathbf{Z}$ as the product of the three orthogonal matrices, which therefore is orthogonal: $\mathbf{Z}'\mathbf{Z}=\mathbf{I}$.
Orthogonality of $\mathbf{Z}$ gives $\forall i$
$$ z_{i1}^2 + z_{i2}^2 + \cdot + z_{ip}^2 = 1 \Rightarrow z_{ii} \ge 1. $$
Hence, (note all singular values are non-negative)
$$ \tr(\bs{\Sigma}’\mathbf{Z}) = \sum_{i=1}^p \sigma_i z_{ii} \le \sum_{i=1}^p \sigma_i $$
which gives optimal $\mathbf{Z}^*=\mathbf{I}$ and thus the solution follows.
2) Orthogonal Procrustes Problem. This seeks the solution to
$$ \min_{\mathbf{X}’\mathbf{X}=\mathbf{I}}\norm{\mathbf{A}-\mathbf{BX}}_F $$
which is, similar to the problem above, given by the SVD of $\mathbf{BA}'=\mathbf{U}\bs{\Sigma} \mathbf{V}'$, namely $\mathbf{X}=\mathbf{UV}'$.
3) Nearest Symmetric Matrix for $\mathbf{A}\in\R^{p\times p}$ seeks solution to $\min\norm{\mathbf{A}-\mathbf{X}}_F$, which is simply
$$ \mathbf{X} = \frac{\mathbf{A}+\mathbf{A}’}{2}. $$
In order to prove it, write $\mathbf{A}$ in the form of
$$ \mathbf{A} = \frac{\mathbf{A} + \mathbf{A}’}{2} + \frac{\mathbf{A} - \mathbf{A}’}{2} =: \mathbf{X} + \mathbf{Y}. $$
Notice $\tr(\mathbf{X}'\mathbf{Y})=0$, hence by Pythagoras we know $\mathbf{Y}$ is the minimum we can find for the problem above.
4) Best Rank-$r$ Approximation. In order to find the best rank-$r$ approximation in Frobenius norm, we need solution to
$$ \min_{\rank(\mathbf{X})\le r} \norm{\mathbf{A}-\mathbf{X}}_F $$
which is merely $\mathbf{X}=\mathbf{U}_r\bs{\Sigma}_r\mathbf{V}_r'$. See condensed SVD above for notation.
The best approximation in 2-norm, namely solution to
$$ \min_{\rank(\mathbf{X})\le r} \norm{\mathbf{A}-\mathbf{X}}_2, $$
is exactly identical to the one above. We may prove both by reduction to absurdity. Proof: Suppose $\exists \mathbf{B}\in\R^{n\times p}$ s.t.
Now choose $\mathbf{w}$ from kernel of $\mathbf{B}$ and we have
$$ \mathbf{Aw}=\mathbf{Aw}+\bs{0} = (\mathbf{A}-\mathbf{B})\mathbf{w} $$
and thus
Meanwhile, note $\mathbf{w}\in\span\{v_1,v_2,\ldots,v_{r+1}\}=\mathbf{W}$, assume particularly $\mathbf{w}=\mathbf{v}_{r+1}\bs{\alpha}$, then
Due to contradiction between eq. (1) and (2) we conclude such $\mathbf{B}$ doesn’t exist.
Orthonormal Bases for Four Subspaces using SVD
SVD can be used to get orthonormal bases for each of the four subspaces: the column space $C(\mathbf{A})$, the null space $N(\mathbf{A})$, the row space $C(\mathbf{A}')$, and the left null space $N(\mathbf{A}')$.
$\mathbf{U}_r$forms a basis of$C(\mathbf{A})$.$\bar{\mathbf{U}}_r$forms a basis of$N(\mathbf{A}')$.$\mathbf{V}_r$forms a basis of$C(\mathbf{A}')$.$\bar{\mathbf{V}}_r$forms a basis of$N(\mathbf{A}')$.
See this post for detailed proof.
Principle Component Analysis (PCA)
PCA is the most important matrix analysis tool. In this section we use $\mathbf{X}=(X_1,X_2,\ldots,X_p)$ to denote a vector of random variables. Being capitalized here means they are random variables rather than observations, and thus a capitalized bold symbol stands still for a vector.
Three Basic Formulas (for Population Analysis)
Expectation:
$$ \E[\mathbf{AX}] = \mathbf{A}\E[\mathbf{X}]. $$
Variance:
$$ \Var[\mathbf{Ax}] = \mathbf{A}\Var[\mathbf{X}]\mathbf{A}’. $$
Covariance:
$$ \Cov[\mathbf{a}’\mathbf{X},\mathbf{b}’\mathbf{X}] = \mathbf{a}’\Var[\mathbf{X}]\mathbf{b}. $$
Definition of Population PCA
Given $\mathbf{X}:\Omega\to\R^p$, find $\mathbf{A}\in\R^{p\times p}$ s.t.
$Y_1,Y_2,\ldots,Y_p$are uncorrelated, where$\mathbf{Y}=(Y_1,Y_2,\ldots,Y_p)=\mathbf{A}\mathbf{X}$.$Y_1,Y_2,\ldots,Y_p$have variances as large as possible.
These two condisions can be described in equations as below:
$\Cov[Y_i,Y_j]=\mathbf{O}$for all$i\neq j$.$\Var[Y_i]$is maximized for all$i=1,2,\ldots,p$(under the restraints that$\norm{\mathbf{a}_j}=1$for all$j=1,2,\ldots,p$).
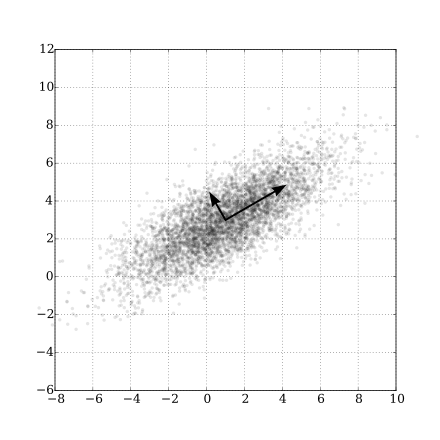
These $Y_1,Y_2,\ldots,Y_p$ are called principle components of $\mathbf{X}$.
How to Calculate Population PCs
We can do it recursively:
- 1st PC: for
$Y_1=\mathbf{a}_1'\mathbf{X}$, let$\mathbf{a}_1=\arg\max\{\Var[\mathbf{a}_1'\mathbf{X}]\}$s.t.$\norm{\mathbf{a}_1}=1$. - 2nd PC: for
$Y_2=\mathbf{a}_2'\mathbf{X}$, let$\mathbf{a}_2=\arg\max\{\Var[\mathbf{a}_2'\mathbf{X}]\}$s.t.$\norm{\mathbf{a}_2}=1$,$\Cov[\mathbf{a}_1'\mathbf{X},\mathbf{a}_2'\mathbf{X}]=0$. - 3rd PC: for
$Y_3=\mathbf{a}_3'\mathbf{X}$, let$\mathbf{a}_3=\arg\max\{\Var[\mathbf{a}_3'\mathbf{X}]\}$s.t.$\norm{\mathbf{a}_3}=1$,$\Cov[\mathbf{a}_1'\mathbf{X},\mathbf{a}_3'\mathbf{X}]=0$and$\Cov[\mathbf{a}_2'\mathbf{X},\mathbf{a}_3'\mathbf{X}]=0$. - …
Or, we can do it analytically by the theorem below:
Theorem Let $\bs{\Sigma}=\Cov[\mathbf{X}]$ and let the EVD be $\bs{\Sigma} = \mathbf{A}\bs{\Lambda}\mathbf{A}'$, then it can be proved that $Y_k = \mathbf{a}_k'\mathbf{X}$ is the $k$-th PC.
Properties of Population PCs
- The total variance is not changed:
$\sum \Var[X_i]=\sum\Var[Y_i]$. - The proportion in variance of the
$k$-th PC is$\frac{\Var[Y_k]}{\sum \Var[Y_i]} = \frac{\lambda_k}{\sum \lambda_i}$where$\lambda_i$is the$i$-th eigenvalue. - The correlation
$\Corr[Y_i, X_j]=\sqrt{\lambda_i}a_{ij}/\sigma_j$.
Definition of Sample PCA
Given $n$ samples: $\{X(\omega_1), X(\omega_2), \ldots, X(\omega_n)\}$. Everything is just the same except being in sample notations, e.g. $\bar{\mathbf{X}}=\mathbf{X}'\bs{1} / n$ and $S=(\mathbf{X}-\bar{\mathbf{X}}'\bs{1})'(\mathbf{X}-\bar{\mathbf{X}}'\bs{1}) / (n-1)$.
How to Calculate Sample PCA
In order to avoid loss of precision in calculating the $\mathbf{S}$, we do SVD instead of EVD, on the mean-centered sample matrix $\mathbf{X}-\bs{1}\bar{\mathbf{X}}=\mathbf{U}\bs{\Sigma} \mathbf{V}'\in\R^{n\times p}$. Then it can be proved that the $k$-th sample PC is given by $\mathbf{v}_k$, $k=1,2,\ldots,p$.
Remarks: In this case, we call $\mathbf{V}$ the loading matrix, and $\mathbf{U}\bs{\Sigma}:=\mathbf{T}$ the score matrix. The PCA scatter plot is thereby the projection onto the PCs, i.e. the columns of $\mathbf{V}$. Specifically, the coordinates are gonna be $\{(t_{ij}, t_{ik})\in\R^2: i=1,2,\ldots, n\}$ namely the selected first columns of the score matrix. This is identical to manually projecting $\mathbf{X}$ onto selected columns of $\mathbf{Q}$ (the principle components) as it can be proved that $t_{ij}=\mathbf{x}_i'\mathbf{q}_j$. However, by using SVD we avoid miss-calculating the eigenvalues in low precision systems.
Definition of Variable PCA
This is merely population PCA on $\mathbf{X}'\in\R^{p\times n}$. Transposing $\mathbf{X}$ swaps the roles of the number of variables and the size of population. The SVD now becomes
$$ \mathbf{X}’ = \mathbf{V}\bs{\Sigma}\mathbf{U}' $$
where we now instead call $\bs{V\Sigma}$ the $\mathbf{T}$ variable.
Remarks: By plotting $\mathbf{X}$ against $\mathbf{V}$ we get PCA scatter plot; by plotting $\mathbf{X}$ against $\mathbf{U}$ on the same piece of paper where draw this PCA scatter plot, we get the so-called biplot.
Application of PCA: Sample Factor Analysis (FA)
One sentence to summarize it: sample factor analysis equals PCA. In formula, FA is trying to write $\mathbf{X}\in\R^p$ into
$$ \mathbf{X} = \bs{\mu} + \mathbf{LF} + \bs{\varepsilon} $$
where $\bs{\mu}\in\R^p$ are $p$ means of features (aka. alphas), $\mathbf{L}\in\R^{p\times m}$ are loadings (aka. betas) and $\mathbf{F}\in\R^m$ are called the factors.
There are some assumptions:
$m\ll p$(describing a lot of features in few factors).$\E[\mathbf{F}] = 0$(means are captured already by$\bs{\mu}$),$\Cov[\mathbf{F}]=\mathbf{I}$(factors are uncorrelated).$\E[\bs{\varepsilon}]=0$(residuals are zero-meaned),$\Cov[\bs{\varepsilon}]=\bs{\Xi}$is diagonal (residuals are uncorrelated).$\Cov[\bs{\varepsilon},\mathbf{F}]=\E[\bs{\varepsilon F}']=\mathbf{O}$($ \bs{\varepsilon}$ is uncorrelated with $\mathbf{F}$).
With these assumptions we have $\bs{\Sigma}=\mathbf{LL}'+\bs{\Xi}$.
Canonical Correlation Analysis (CCA)
Similar to PCA, we try to introduce CCA in two ways, namely w.r.t. a population and a sample.
Notations for Population CCA
Assume $p\le q$ (important!!!). Given $\mathbf{X}\in\R^p$ and $\mathbf{Y}\in\R^q$, define
and
Furthermore, define
and
Also, let
Then with this notation we know given $\mathbf{a}\in\R^p$ and $\mathbf{b}\in\R^q$, how expectation, variance and covariance (ans thus correlation) are represented for $U=\mathbf{a}'\mathbf{X}$ and $V=\mathbf{b}'\mathbf{Y}$.
Definition of Population CCA
We calculate canonical correlation variables iteratively:
$(U_1,V_1)=\arg\max\{\Cov[U,V]:\Var[U]=\Var[V]=1\}$.$(U_2,V_2)=\arg\max\{\Cov[U,V]:\Var[U]=\Var[V]=1,\Cov[U,U_1]=\Cov[V,V_1]=\Cov[V,U_1]=\Cov[U,V_1]=0\}$.- …
$(U_k,V_k)=\arg\max\{\Cov[U,V]:\Var[U]=\Var[V]=1,\Cov[U,U_i]=\Cov[V,V_i]=\Cov[V,U_i]=\Cov[U,V_i]=0\ \ \forall i=1,2,\ldots, k-1\}$.
Theorem Suppose $p\le q$, let $\Gamma_{\mathbf{XY}}=\bs{\Sigma}_{\mathbf{X}}^{-1/2}\bs{\Sigma}_{\mathbf{XY}}\bs{\Sigma}_{\mathbf{Y}}^{-1/2}\in\R^{p\times q}$ and the condensed SVD be1
$$ \bs{\Gamma}_{\mathbf{XY}} = \begin{bmatrix} \mathbf{u}_1 & \mathbf{u}_2, & \ldots & \mathbf{u}_p \end{bmatrix}\begin{bmatrix} \sigma_1 & \cdots & \cdots & \mathbf{O} \ \vdots & \sigma_2 & \cdots & \vdots \ \vdots & \vdots & \ddots & \vdots \ \mathbf{O} & \cdots & \cdots & \sigma_p \end{bmatrix}\begin{bmatrix} \mathbf{v}_1’\ \mathbf{v}_2’ \ \vdots \ \mathbf{v}_p' \end{bmatrix} $$
which gives
and that $\rho_k=\sigma_k$.
We call $\rho_k=\Corr[U_k,V_k]$ as the $k$-th population canonical correlation. We call $\mathbf{a}_k=\bs{\Sigma}_{\mathbf{X}}^{-1/2}\mathbf{u}_k$ and $\mathbf{b}_k=\bs{\Sigma}_{\mathbf{Y}}^{-1/2}\mathbf{v}_k$ as the population canonical vectors.
Properties of Population CCA
The canonical correlation variables have the following three basic properties:
$\Cov[U_i,U_j]=\1{i=j}$.$\Cov[V_i,V_j]=\1{i=j}$.$\Cov[U_i,V_j]=\1{i=j} \sigma_i$.
Theorem Let $\tilde{\mathbf{X}}=\mathbf{MX}+\mathbf{c}$ be the affine transformation of $\mathbf{X}$. Similarly let $\tilde{\mathbf{Y}}=\mathbf{NY}+\mathbf{d}$. Then by using CCA, the results from analyzing $(\tilde{\mathbf{X}},\tilde{\mathbf{Y}})$ remains unchanged as $(\mathbf{X},\mathbf{Y})$, namely CCA is affine invariant.
Based on this theorem we have the following properties:
- The canonical correlations between
$\tilde{\mathbf{X}}$and$\tilde{\mathbf{Y}}$are identical to those between$\mathbf{X}$and$\mathbf{Y}$. - The canonical correlation vectors are not the same. They now becomes
$\tilde{\mathbf{a}_k}=(\mathbf{M}')^{-1}\mathbf{a}_k$and$\tilde{\mathbf{b}_k}=(\mathbf{N}')^{-1}\mathbf{b}_k$. - By using covariances
$\bs{\Sigma}$or correlations$\mathbf{P}$makes no difference in CCA2. This is not true for PCA, i.e. there is no simple relationship between PCA obtained from covariances and PCA from correlations.
Example of Calculating Population CCA
Let’s assume $p=q=2$, let
with $|\alpha| < 1$, $|\gamma| < 1$. In order to find the 1st canonical correction of $\mathbf{X}$ and $\mathbf{Y}$, we first check
Therefore, we may calculate
It’s easy to show that $\lambda_1= 2$ and $\lambda_2=0$ are the two eigenvalues of $\bs{11}'$ and thus the 1st canonical correction is
$$ \rho_1 = \sqrt{\frac{4\beta^2}{(1+\alpha)(1+\gamma)}} = \frac{2\beta}{\sqrt{(1+\alpha)(1+\gamma)}}. $$
Notations for Sample CCA
Given $n$ samples: $\mathbf{X}=\{\mathbf{X}(\omega_1), \mathbf{X}(\omega_2), \ldots, \mathbf{X}(\omega_n)\}\in\R^{n\times p}$ and similarly $\mathbf{Y}\in\R^{n\times q}$. Everything is just the same except being in sample notations, e.g. $\bar{\mathbf{X}}=\mathbf{X}'\bs{1} / n$ and $S_{\mathbf{X}}=(\mathbf{X}-\bar{\mathbf{X}}'\bs{1})'(\mathbf{X}-\bar{\mathbf{X}}'\bs{1}) / (n-1)$.
Besides the regular notations, here we also define $r_{\mathbf{XY}}(\mathbf{a},\mathbf{b})$ as the sample correlation of $\mathbf{a}'\mathbf{X}$ and $\mathbf{b}'\mathbf{Y}$:
Definition of Sample CCA
Same as population CCA, we give sample CCA iteratively (except that here we’re talking about canonical correlation vectors directly):
$(\hat{\mathbf{a}}_1,\hat{\mathbf{b}}_1)=\arg\max\{\mathbf{a}'\mathbf{S}_{\mathbf{XY}}\mathbf{b}:\mathbf{a}'\mathbf{S}_{\mathbf{X}}\mathbf{a}=\mathbf{b}'\mathbf{S}_{\mathbf{Y}}\mathbf{b}=1\}$.$(\hat{\mathbf{a}}_2,\hat{\mathbf{b}}_2)=\arg\max\{\mathbf{a}'\mathbf{S}_{\mathbf{XY}}\mathbf{b}:\mathbf{a}'\mathbf{S}_{\mathbf{X}}\mathbf{a}=\mathbf{b}'\mathbf{S}_{\mathbf{Y}}\mathbf{b}=1,\mathbf{a}'\mathbf{S}_{X}\hat{\mathbf{a}}_1=\mathbf{b}'\mathbf{S}_{\mathbf{Y}}\hat{\mathbf{b}}_1=\mathbf{a}'\mathbf{S}_{\mathbf{XY}}\hat{\mathbf{b}}_1=\mathbf{b}'\mathbf{S}_{\mathbf{YX}}\hat{\mathbf{a}}_1=0\}$.- …
$(\hat{\mathbf{a}}_k,\hat{\mathbf{b}}_k)=\arg\max\{\mathbf{a}'\mathbf{S}_{\mathbf{XY}}\mathbf{b}:\mathbf{a}'\mathbf{S}_{\mathbf{X}}\mathbf{a}=\mathbf{b}'\mathbf{S}_{\mathbf{Y}}\mathbf{b}=1,\mathbf{a}'\mathbf{S}_{X}\hat{\mathbf{a}}_i=\mathbf{b}'\mathbf{S}_{\mathbf{Y}}\hat{\mathbf{b}}_i=\mathbf{a}'\mathbf{S}_{\mathbf{XY}}\hat{\mathbf{b}}_i=\mathbf{b}'\mathbf{S}_{\mathbf{YX}}\hat{\mathbf{a}}_i=0\}\ \ \forall i=1,2,\ldots,k-1$.
Theorem Given $p\le q$, let $\mathbf{G}_{\mathbf{XY}} = \mathbf{S}_{\mathbf{X}}^{-1/2}\mathbf{S}_{\mathbf{XY}}\mathbf{S}_{\mathbf{Y}}^{-1/2}\in\R^{p\times q}$ and the SVD be
Then we have
In addition,
$$ r_k = r_{\mathbf{XY}}(\hat{\mathbf{a}}_k, \hat{\mathbf{b}}_k) = \sigma_k. $$
We call $r_k$ the $k$-th sample canonical correlation. Also, $\mathbf{Xa}_k$ and $\mathbf{Yb}_k$ are called the score vectors.
Properties of Sample CCA
Everything with population CCA, including the affine invariance, holds with sample CCA, too.
Example of Calculating Sample CCA
Let $p=q=2$, let
$$ \mathbf{X}=\begin{bmatrix} \text{head length of son1}\ \text{head breath of son1} \end{bmatrix}=\begin{bmatrix} l_1\ b_1 \end{bmatrix}\quad\text{and}\quad \mathbf{Y}=\begin{bmatrix} \text{head length of son2}\ \text{head breath of son2} \end{bmatrix}=\begin{bmatrix} l_2\ b_2 \end{bmatrix}. $$
Assume there’re $25$ families, each having $2$ sons. The $\mathbf{W}$ matrix is therefore
$$ \mathbf{W} = \begin{bmatrix} \mathbf{l}_1 & \mathbf{b}_1 & \mathbf{l}_2 & \mathbf{b}_2 \end{bmatrix}\in\R^{25\times 4}. $$
In addition, we may also calculate the correlations $\mathbf{R}_{\mathbf{X}}$, $\mathbf{R}_{\mathbf{Y}}$ and $\mathbf{R}_{\mathbf{XY}}$, from which we have $\mathbf{G}_{\mathbf{XY}}$ given by
which gives the sample canonical correlations $r_1$ and $r_2$ (in most cases $r_1\ll r_2$). In the meantime, we have $\mathbf{u}_{1,2}$ and $\mathbf{v}_{1,2}$ and because of significant difference in scale of $r_1$ and $r_2$, we don’t care about $\mathbf{u}_2$ and $\mathbf{v}_2$. From $\mathbf{u}_1$ and $\mathbf{v}_1$ we can calculate $\hat{\mathbf{a}}_1$ and $\hat{\mathbf{b}}_2$, which indicate the linear relationship between the features. Specifically, the high correlation $r_1$ tells that $\hat{U}_1=\hat{\mathbf{a}}_1\mathbf{X}$ and $\hat{V}_1=\hat{\mathbf{b}}_1\mathbf{Y}$, which are essentially the “girths” (height $+$ breath) of sons’ faces, are highly correlated. In comparison, data shows that $\hat{U}_2$ and $\hat{V}_2$, which describes the “shapes” (height $-$ breath) of sons’ faces, are poorly correlated.
Linear Discriminant Analysis (LDA)
Different from the unsupervised learning algorithms we’ve been discussing in the previous sections, like PCA, FA and CCA, LDA is a supervised classification method in that the number of classes to be divided into is specified explicitly.
Notations for LDA
Assume $g$ different classes $C_1,C_2,\ldots,C_g$. We try to solve the problem asking for the optimal division of $n$ rows of $\mathbf{X}\in\R^{n\times p}$ into $g$ parts: for each $i=1,2,\ldots,g$ denote
with $\sum_{i=1}^g n_i=g$, then given $\mathbf{a}\in\R^p$ we can define
$$ \mathbf{X}_i\mathbf{a} =: \mathbf{y}_i \in \R^{n_i},\quad i=1,2,\ldots,g. $$
Now, recall the mean-centering matrix
$$ \mathbf{H} = \mathbf{I} - \frac{\bs{\iota}\bs{\iota}’}{\bs{\iota}’\bs{\iota}} $$
which provides handy feature that centers a vector by its mean: for any $\mathbf{X}$
$$ \mathbf{HX} = \mathbf{X} - \bs{\iota}’\bar{\mathbf{X}}. $$
With $\mathbf{H}$ we also have the total sum-of-squares as
$$ \mathbf{T} = \mathbf{X}’\mathbf{HX} = \mathbf{X}’\left(\mathbf{I}-\frac{\bs{\iota}\bs{\iota}’}{\bs{\iota}’\bs{\iota}}\right)\mathbf{X} = (n-1)\mathbf{S}. $$
Similarly we define $\mathbf{H}_i$ and $\mathbf{W}_i=\mathbf{X}_i'\mathbf{H}_i\mathbf{X}_i$ for each $i=1,2,\ldots,g$ and thus
where $\mathbf{W}\in\R^{p\times p}$ is known as the within-group sum-of-squares. Finally, check
where we define $\mathbf{B}\in\R^{p\times p}$ as the between-group sum-of-squares.
Theorem For any $\mathbf{a}\in\R^p$, $\mathbf{a}'\mathbf{T}\mathbf{a} = \mathbf{a}'\mathbf{B}\mathbf{a} + \mathbf{a}'\mathbf{W}\mathbf{a}$. However, $\mathbf{T}\neq \mathbf{B}+\mathbf{W}$.
Definition of LDA
First let’s give the Fisher Linear Discriminant Function
$$ f(\mathbf{a}) = \frac{\mathbf{a}’\mathbf{B}\mathbf{a}}{\mathbf{a}’\mathbf{W}\mathbf{a}} $$
by maximizing which, Fisher states, gives the optimal classification.
Theorem Suppose $\mathbf{W}\in\R^{p\times p}$ is nonsingular, let $\mathbf{q}_1\in\R^p$ be the principle eigenvector of $\mathbf{W}^{-1}\mathbf{B}$ corresponding to $\lambda_1=\lambda_{\max}(\mathbf{W}^{-1}\mathbf{B})$, then it can be shown that $\mathbf{q}_1=\arg\max_{\mathbf{a}}f(\mathbf{a})$ and $\lambda_1=\max_{\mathbf{a}}f(\mathbf{a})$.
Proof. We know
$$ \max_{\mathbf{a}} \frac{\mathbf{a}’\mathbf{B}\mathbf{a}}{\mathbf{a}’\mathbf{W}\mathbf{a}} = \max_{\mathbf{a}’\mathbf{W}\mathbf{a}=1} \mathbf{a}’\mathbf{B}\mathbf{a} = \max_{\mathbf{b}’\mathbf{b}=1} \mathbf{b}’\mathbf{W}^{-1/2}\mathbf{B}\mathbf{W}^{-1/2}\mathbf{b} = \lambda_{\max}(\mathbf{W}^{-1/2}\mathbf{B}\mathbf{W}^{-1/2}) = \lambda_{\max}(\mathbf{W}^{-1}\mathbf{B}). $$
Therefore, we have the maximum $\lambda_1$ and thereby we find the maxima $\mathbf{q}_1$.Q.E.D.
Remark: $\lambda_1$ and $\mathbf{q}_1$ can also be seen as the solutions to
$$ \mathbf{B}\mathbf{x} = \lambda \mathbf{W}\mathbf{x},\quad \mathbf{x}\neq\bs{0} $$
which is known as a generalized eigenvalue problem since it reduces to the usual case when $\mathbf{W}=\mathbf{I}$.
Classification Rule of LDA
Given a new data point $\mathbf{t}\in\R^p$ we find
$$ i = \underset{j=1,2,\ldots,g}{\arg\min} |\mathbf{q}_1’(\mathbf{t}-\bar{\mathbf{X}_j})| $$
and assign $\mathbf{t}$ to class $C_j$. This essentially the heuristic behind Fisher’s LDA.
There is no general formulae for $g$-class LDA, but we do have one for specifically $g=2$.
Population LDA for Binary Classification
The population linear discriminant function is
$$ f(\mathbf{a}) = \frac{\sigma_{\text{between}}}{\sigma_{\text{within}}} = \frac{(\mathbf{a}’\bs{\mu}_1 - \mathbf{a}’\bs{\mu}-2)^2}{\mathbf{a}’\bs{\Sigma}_1\mathbf{a} + \mathbf{a}’\bs{\Sigma}_2\mathbf{a}} = \frac{[\mathbf{a}’(\bs{\mu}_1-\bs{\mu}_2)]^2}{\mathbf{a}’(\bs{\Sigma}_1+\bs{\Sigma}_2)\mathbf{a}} $$
which solves to
$$ \mathbf{q}_1 = (\bs{\Sigma}_1 + \bs{\Sigma}_2)^{-1}(\bs{\mu}_1-\bs{\mu}_2) := \bs{\Sigma}^{-1}(\bs{\mu}_1-\bs{\mu}_2) $$
and the threshold
$$ c=\frac{1}{2}(\bs{\mu}_1’\bs{\Sigma}^{-1}\bs{\mu}_1 - \bs{\mu}_2’\bs{\Sigma}^{-1}\bs{\mu}_2). $$
The classification is therefore given by
$$ C(\mathbf{t}) = \begin{cases} C_1\ & \text{if}\quad\mathbf{q}_1’\mathbf{t} > c,\ C_2\ & \text{if}\quad\mathbf{q}_1’\mathbf{t} < c.\ \end{cases} $$
MDS and CA
Variable matrix decomposition methods provide different classes of data analysis tools:
- Based on SVD we have PCA, FA, HITS and LSI for general
$\mathbf{A}\in\R^{n\times p}$. - Based on EVD we have CCA and MDS for
$\mathbf{A}\in\R^{p\times p}$, symmetric. - Based on generalized EVD (GEVD) we have LDA for
$\mathbf{A},\mathbf{B}\in\R^{p\times p}$, both symmetric. - Finally, based on generalized SVD (GSVD) we have CA for
$\mathbf{A}=\mathbf{U}\bs{\Sigma}\mathbf{V}'\in\R^{n\times p}$where we instead of orthonormality of$\mathbf{U}$and$\mathbf{V}$assume$\mathbf{U}'\mathbf{D}_1\mathbf{U}=\mathbf{I}$and$\mathbf{V}'\mathbf{D}_2\mathbf{V}=\mathbf{I}$for diagonal$\mathbf{D}_1\in\R^{n\times n}$and$\mathbf{D}_2\in\R^{p\times p}$.
-
Every symmetric positive definite matrix has a square root given by
$\mathbf{A}^{1/2} = \mathbf{Q}\bs{\Lambda}^{1/2}\mathbf{Q}'$where$\mathbf{A}=\mathbf{Q}\bs{\Lambda}\mathbf{Q}'$is the EVD of$\mathbf{A}$. ↩︎ -
In fact the canonical correlation vectors are scaled by
$\mathbf{V}^{-1/2}_{\mathbf{X}}$and$\mathbf{V}^{-1/2}_{\mathbf{Y}}$respectively. ↩︎
